Universal Proteomics Resources
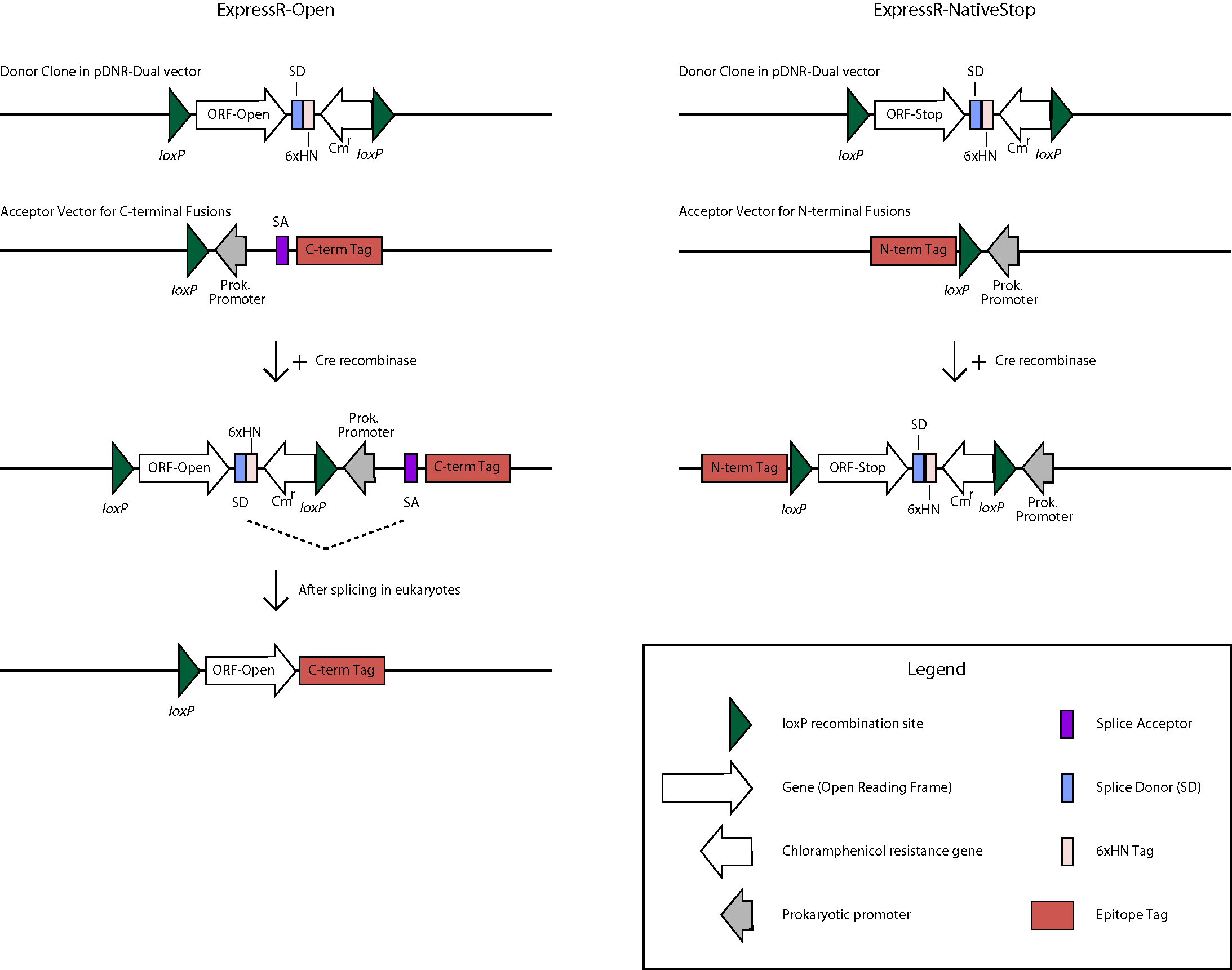
The BDGP is in the process of transferring over 7,000 high-quality ORFs into a universal donor vector. The ORFs originate from clones in our Gold Collection which serves as the primary resource for high-throughput proteomics applications. This is being accomplished with the use of the Clontech In-Fusion PCR cloning system (view Cloning Protocol) and the donor vector pDNR-Dual. In order to make the universal donor clone collection as useful as possible, two forms of each ORF are being made- one containing the native stop codon and one that lacks the stop codon and are referred to as the ExpressR-NativeStop(NS) and ExpressR-Open(O) clone collections, respectively. For expression, transfer of ORFs is mediated by in-vitro Cre-lox recombination to appropriate expression vectors (see below). The NS and O forms of the clones will allow for amino-terminal or carboxy-terminal fusion proteins, respectively. Diagrams of the directional cloning process flow and the bioinformatics process illustrate the subcloning process and the database steps and software used to track our progress and evaluate quality.
 Click for larger image Click here to see
nucleotide sequences for selected features from the pDNR-Dual and acceptor vectors (i.e. Splice Donor, Splice
Acceptor, loxP)
Click for larger image Click here to see
nucleotide sequences for selected features from the pDNR-Dual and acceptor vectors (i.e. Splice Donor, Splice
Acceptor, loxP)
Search for Expression Clones
Universal Donor Clone Collection
Selected clones from the ExpressR-NS (clones beginning with a "BS" prefix) and ExpressR-O (clones beginning with a "BO" prefix) clone collections are full-length sequenced and compared against the FlyBase protein translations. Clones that match the amino-acid translation 100% along the entire length of the ORF are colony-purified and re-arrayed into plates with an "XS" (native-stop) or "XO" (open) prefix. These plates are then end-sequenced for clone verification and are available from the [Drosophila Genomic Resources Center at Indiana University, Bloomington](https://dgrc.cgb.indiana.edu/). As the collections grow, we will periodically ship plates to distributors and make announcements on the BDGP web site. The contents of the initial plates of Transcription Factor clones can be [downloaded](/EST/XOXS_TF_details.html). The complete list of all ExpressR-NS and ExpressR-O clones is also available [for download](/EST/XOXS_details.html). [an error occurred while processing this directive]Expression Clone Collections and Vectors
Once plates of the ExpressR-NS and ExpressR-O clones have been generated, the next step is to move the ORFs into appropriate expression vectors. This is accomplished using Cre-lox site-specific recombination, which transfers the ORF from the Donor clone to an Acceptor vector. We have converted four widely used [expression vectors](/about/methods/BDExpression_vectors.html) to accept ORFs in this way. The complete list of clones is also available [for download](/EST/expression_clone_details.html). [an error occurred while processing this directive]Expression Clone Sequence Generator
Use the form below to generate predicted nucleotide sequences and maps of the expression clones.N-terminal Fusion Constructs
Translation for N-terminal fusion proteins will begin at the first methionine (ATG) Start Codon of the N-terminal tag, then proceed in-frame through the loxP, continue through the ORF, and terminate at the native Stop Codon of the ORF. The sequence length between the N-terminal tag and the loxP site may vary by vector but the frame will be maintained. The 6xHN tag will not be expressed.
The structure will appear as follows: Tag-Linker-ORF
C-terminal Fusion Constructs
Translation for C-terminal fusion proteins will begin at the first methionine (ATG) Start Codon of the ORF, then proceed in-frame to the Splice Donor (SD) site.
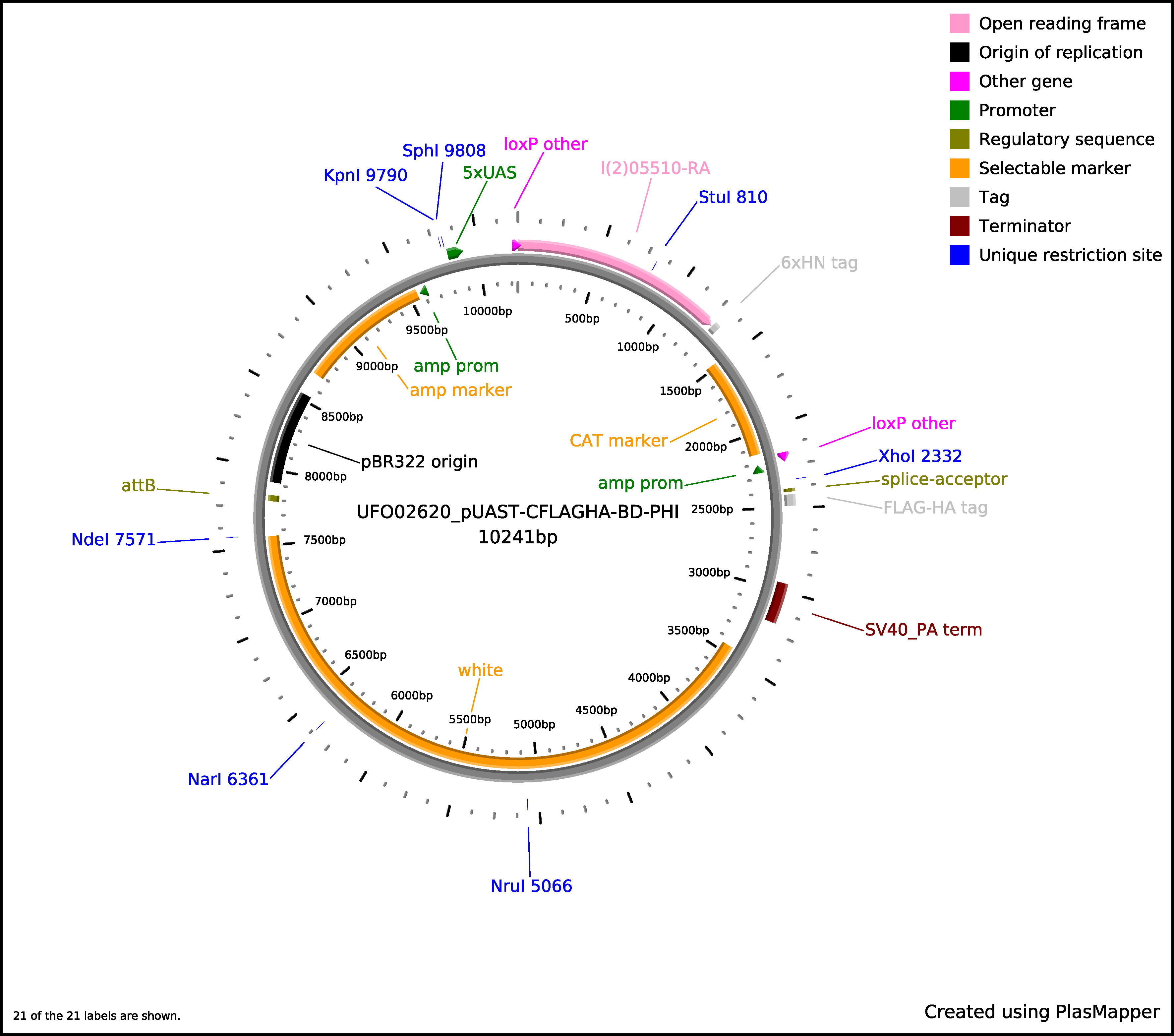
For prokaryotic expression, translation will continue through the 6xHN tag and terminate at the stop codon. For eukaryotic expression, splicing will occur between the SD site and the Splice Acceptor (SA) site, continue through the linker and C-terminal tag, and terminate at the Stop Codon of the tag. The sequence length between the SA site and the C-terminal tag may vary by vector but the frame will be maintained. The 6xHN tag will not be expressed. As an example, here is the predicted protein sequence for clone UFO02620, which was made from clone BO07925 and cloned into the pUAST-CFLAGHA-BD-PHI vector: qsm-PA
Legend: Linker ORF FLAG tag HA tag
MLLSMQMWRSLWLAALFCGLAQAKGSHKVHCSEDQMRVDIGLPDAESKDQS APQIYLEGLKGYPDERCQPQIDGSLAVFRLSLSDFYECGVTRMVNQLTGK KVYYHKIIIESTSGKEIVSVKCITTASPAYNVMMNATTGSSSTSTSSGGI HGLVKRDVLPAGFQEPEDLEITTSLTKRAPEPRLSIGVSQDGQKFTRDLT VKSGTPLTMEINLDEDSAPVYGLGVNYLDVTDTHTSSETLIFKGCTVDPY LFENFNTIDGDILSAKFKAFKFPDSSYVQFRATVNVCLDKCLGTQCSNNQ VGFGRRKREISSANKVYEISLAMFLQVQDIEGVNKNEVLQLEEKLRELKL ANQRLARNSRGNFAMEQTPASAQPAFVVDERELGHLSAGSGAASNGLSLA LWTILGAWSWRLMASFLDHSFGARAQVS >> >> LPVASRDYKDDDDK VKLYPYDVPDYAAA Splice junction is indicated by >> >>.
The estimated size is 50KD.
Transcription Factor Collection
In an effort to make functionally distinct collections of our cDNAs, we are in the process of generating a collection of full-length Transcription Factor clones for use in functional studies. There are an estimated 746 genes in Drosophila that are broadly classified as "Transcription Factors". We have been able to isolate a full-length cDNA for 433 of these factors and we are currently moving their ORFs into pDNR-Dual as described above. A list of clones currently in progress can be [downloaded](/EST/TFweblist433.html).In addition we have also made 36 sequenced-verified transcription factor clones using the Gateway system. These clones, in collaboration with Orbigen, Inc., have been transferred into 2 Baculovirus expression constructs: one that carries an N-terminal 6xHIS fusion and another with an N-terminal TEV-cleavable GST fusion. High-titer stocks of these clones are available at Orbigen, Inc

 Click for larger
image
Click for larger
image